As the volumes of data produced globally continue to surge, so do attempts to find more exotic ways of storing it at lower physical scale than magnetic tape, hard disk drives, or flash storage. Now Microsoft researchers – who have previously demonstrated the use of ultrafast laser optics to store data in quartz glass – say that they have made a major breakthrough that supports storing data on strands of synthetic DNA; the experiment, they say, is “proof that nanoscale DNA writing is possible at dimensions necessary for practical DNA data storage.”
By demonstrating improved control over the "electronic-to-molecular interface" the research success also opens up the possibility of new techniques for drug discovery, tools for assays that detect disease biomarkers or even a platform for sensing environmental pollutants, lead author and Microsoft research director Karin Strauss -- also an Affiliate Professor at the School of Computer Science and Engineering at University of Washington -- claimed.
Experiments on practical DNA data storage have been happening for some years – with proposals for the method spanning both “living vector” organisms like bacteria and the use of synthesised DNA. (Issues with the former, as one pioneer, Nick Goldman notes in a 2017 patent application, include the risks that “germline and somatic mutation will cause the fidelity of stored information and decoded information to be reduced over time.”)
See also: CIO Ian Crawford on exploding film canisters, archiving PBs of data, and Peter Jackson.
The idea, crudely*, involves translating the ones and zeros of binary code into strands of synthetic DNA representing these bits with encoding software and a DNA synthesizer. Why? As Mark Bathe, an MIT professor of biological engineering earlier put it: “DNA is a thousandfold denser than even flash memory, and another property that’s interesting is that once you make the DNA polymer, it doesn’t consume any energy.”
(Citing IDC research that suggests demand for data storage will hit 9 zettabytes a year by 2024, Microsoft’s researchers note that it will take millions of tape cartridges—the current densest commercial storage media—to store 9 zettabytes of information, but “the footprint of one small refrigerator if stored in DNA”.)
Practical DNA data storage: Tackling DNA write density
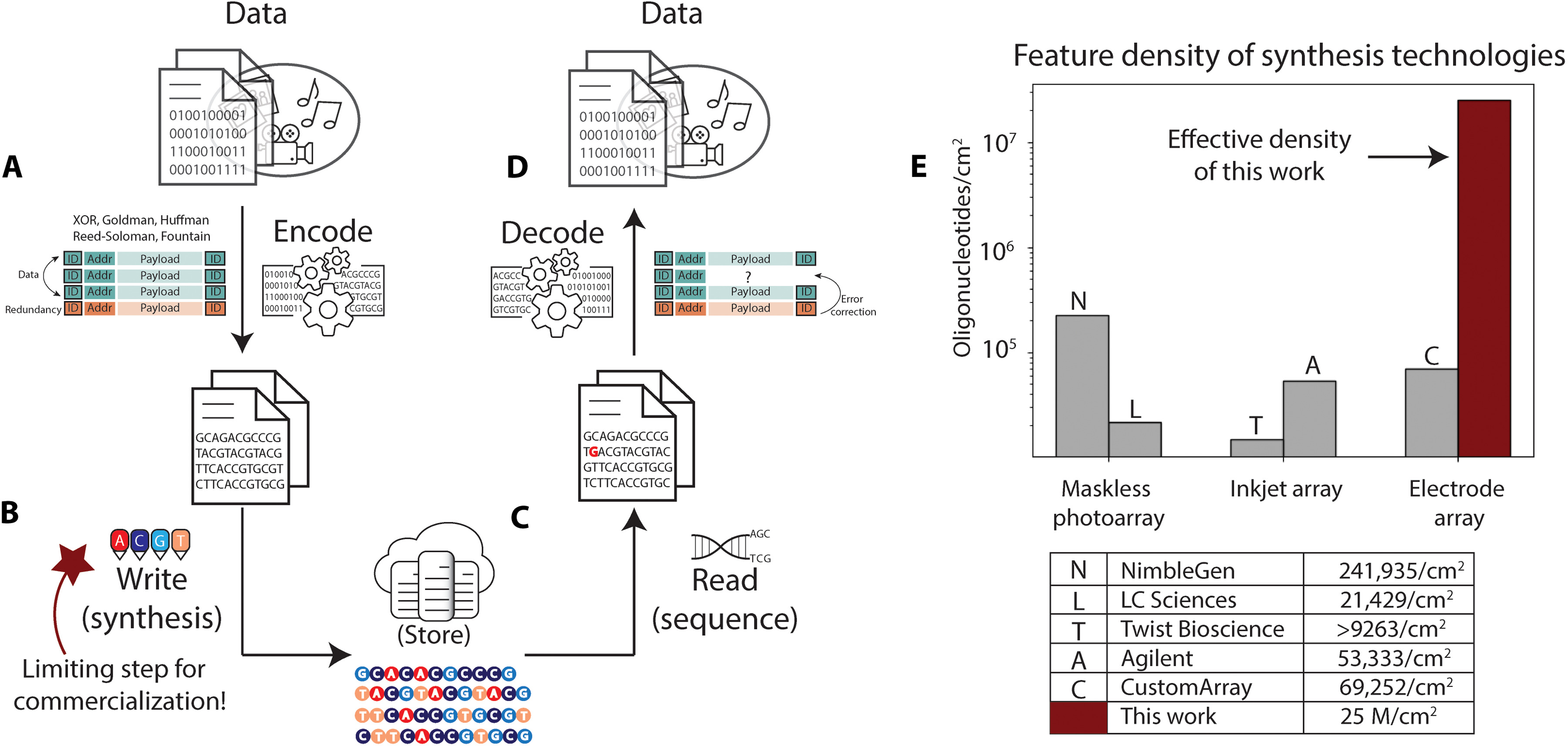
Now the new research from Microsoft, detailed in a November 24, 2021 paper published in Science Advances, “Scaling DNA Data Storage with Nanoscale Electrode Wells, suggests that the company has made a major step towards tackled the “most challenging hurdle in deployment of DNA data storage… the write throughput.”
“We have developed the first nanoscale DNA storage writer, which we expect to scale DNA write density to 25 × 106 sequences per square centimeter, three orders of magnitude improvement over existing DNA synthesis arrays” authors Karin Strauss and Bichlien Nguyen wrote, alongside their 14 co-authors (listed below**).
“Current DNA synthesis arrays are designed for generating a small number of high-quality DNA sequences with millions of exact copies and are achieved through three main array synthesis methods: photochemistry, fluid deposition, and electrochemistry” they note. The latter, they argue, offers the most potential can and the ability to “leverage the semiconductor roadmap where 7 nanometer (nm) feature sizes are common.”
"We already know that DNA is a robust way to store information because we can extract it from bones of woolly mammoths, which date back tens of thousands of years, and still make sense of it" -- Nick Goldman, European Bioinformatics Institute, Cambridge, UK
Using electrochemical DNA synthesis, the team produced an electrode array that, they wrote, "demonstrated independent electrode-specific control of DNA synthesis with electrode sizes and pitches that enable synthesis density of 25 million oligonucleotides/cm2, the estimated electrode density required to achieve the minimum target of kilobytes per second of data storage in DNA" in work that they claimed on December 1 "pushes the state of the art in electronic-chemical control, outpaces the previously reported densest synthesis of arbitrary DNA sequences by a margin of three orders of magnitude, and provides the first experimental indication that the write bandwidth required for DNA data storage can be achieved". (Read the research paper in full here.)
The breakthrough comes as the rise of DNA as a potential storage medium has been fueled by progress in parallelized DNA synthesis and DNA sequencing technologies, such as portable sequencing devices, although cost, stability, latency and other issues remain. The attraction of its use for storage is clear however.
As Barry L. Merriman, Tim Geiser and Paul Mola of Roswell Technologies noted in another patent application in 2017: "What makes DNA attractive for information storage is the extremely high information density resulting from molecular scale storage of information. In theory for example, all human-produced digital information recorded to date, estimated [at the time] to be approximately 1 ZB (ZettaByte) (˜1021 Bytes), could be recorded in less than 1022 DNA bases, or 1/60th of a mole of DNA bases, which would have a mass of just 10 grams."
As Strauss noted this month meanwhile: "Our proof of concept paves a road toward generating massive numbers of unique DNA sequences in parallel for data storage. By injecting electrons at specific locations, we can control the molecular environment surrounding the electrodes and thus control the sequence of the DNA grown there. A natural next step is to embed digital logic in the chip to allow individual control of millions of electrode spots to write kilobytes per second of data in DNA. From there, we foresee the technology reaching arrays containing billions of electrodes capable of storing megabytes per second of data in DNA.
"This will bring DNA data storage performance and cost significantly closer to tape. We welcome further discussions to fully realize more widely available molecular controllers in the future.
Follow The Stack on LinkedIn
*A more detailed description: “Sequences of bits are typically encoded in sequences of the four natural DNA bases. These are “written” into molecular form through de novo DNA oligonucleotide synthesis, which creates specified molecules via a set of repeating chemical steps using phosphoramidite chemistry. Once synthesised, the resulting oligonucleotides are preserved and stored. When data need to be accessed, the DNA storing it is selectively amplified using polymerase chain reaction and sequenced, returning the DNA base sequences to the digital domain. The DNA base sequences are decoded to recover the original sequence of bits.”)
**Bichlien Nguyen, Christopher Takahashi, Gagan Gupta, Jake Smith, Richard Rouse, Paul Berndt, Sergey Yekhanin, David Ward, Siena Ang, Patrick Garvan, Hsing-Yeh Parker, Rob Carlson, Douglas Carmean, Luis Ceze, Karin Strauss









